GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.
Python package for analysis of multiomic single cell RNA-seq and ATAC-seq. - GitHub - cistrome/MIRA: Python package for analysis of multiomic single cell RNA-seq and ATAC-seq.
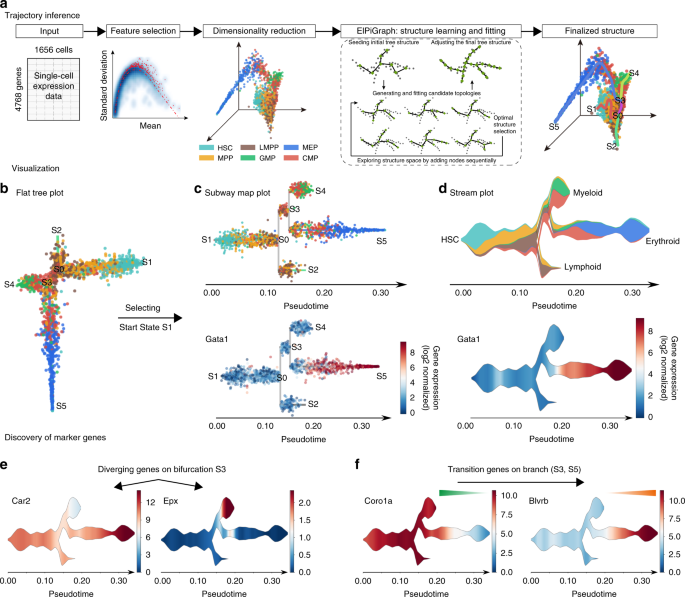
Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM

sciCAN: single-cell chromatin accessibility and gene expression data integration via cycle-consistent adversarial network

MIRA: Joint regulatory modeling of multimodal expression and chromatin accessibility in single cells
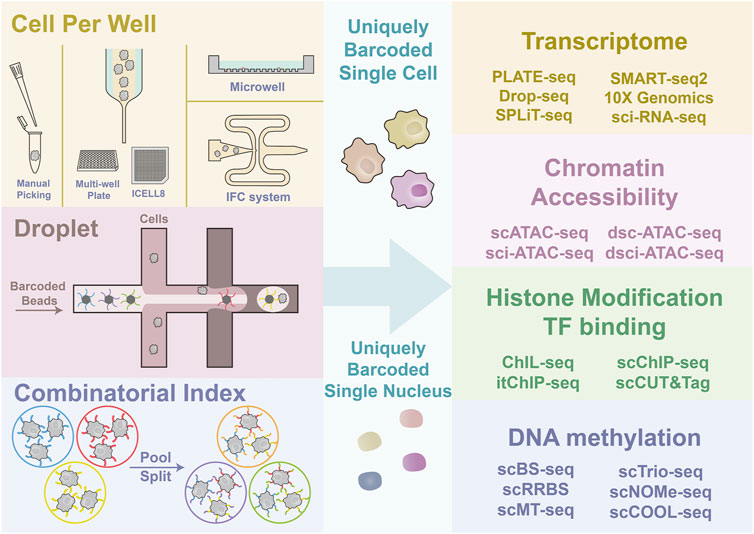
Frontiers Advances in Single-Cell Multi-Omics and Application in Cardiovascular Research

Processing single-cell RNA-seq data for dimension reduction-based analyses using open-source tools - ScienceDirect

Diversification of reprogramming trajectories revealed by parallel single- cell transcriptome and chromatin accessibility sequencing

Single-cell transcriptomic analysis of mIHC images via antigen mapping
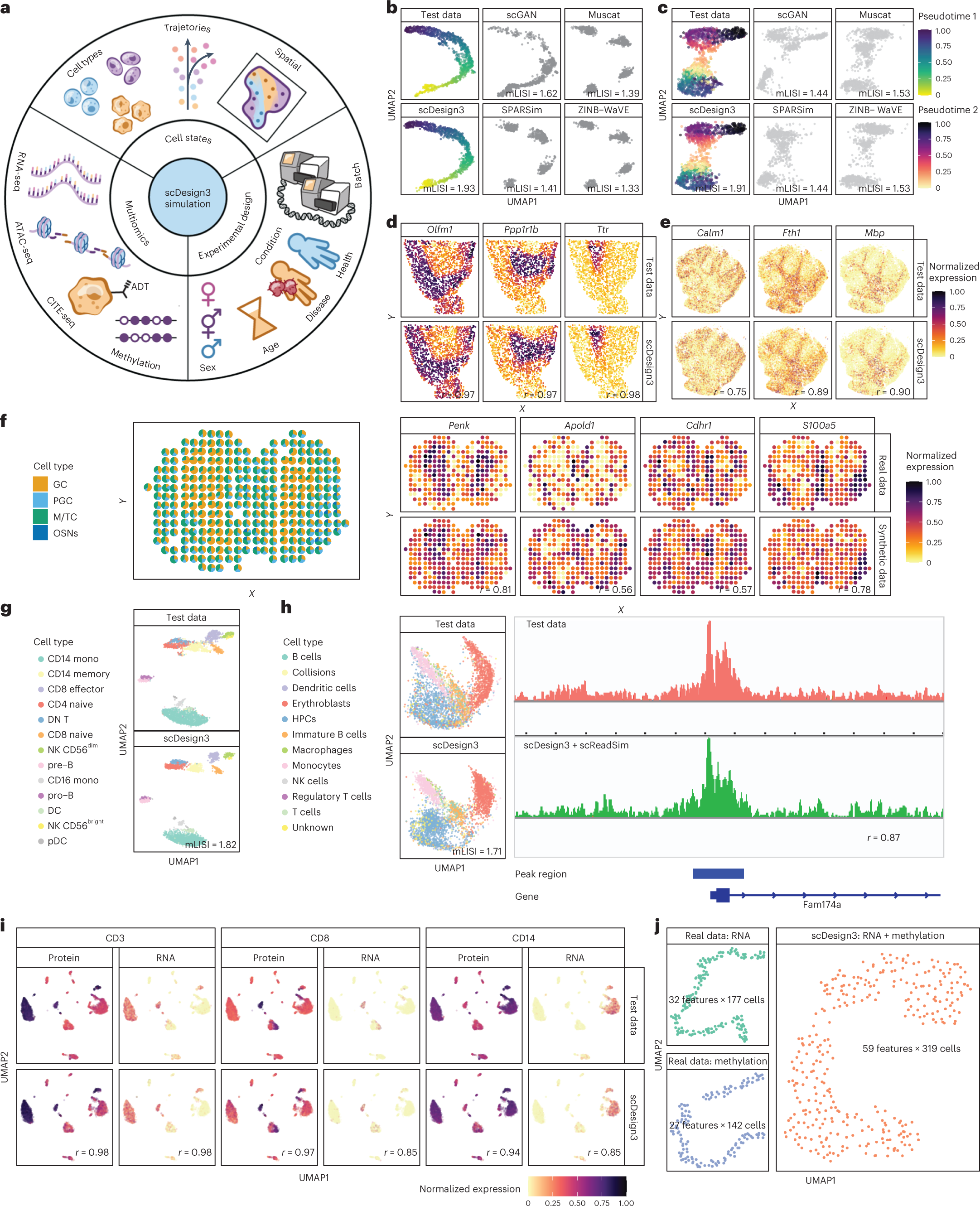
scDesign3 generates realistic in silico data for multimodal single-cell and spatial omics

Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia
ctheodoris · GitHub

APEC: an accesson-based method for single-cell chromatin accessibility analysis, Genome Biology
RNA-seq_notes/README.md at master · mdozmorov/RNA-seq_notes · GitHub

Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells

Kejie Li (@likejie) / X
sc-atacseq-explorer/sc-atacseq-explorer.ipynb at master · JetBrains-Research/sc-atacseq-explorer · GitHub








